Introduction
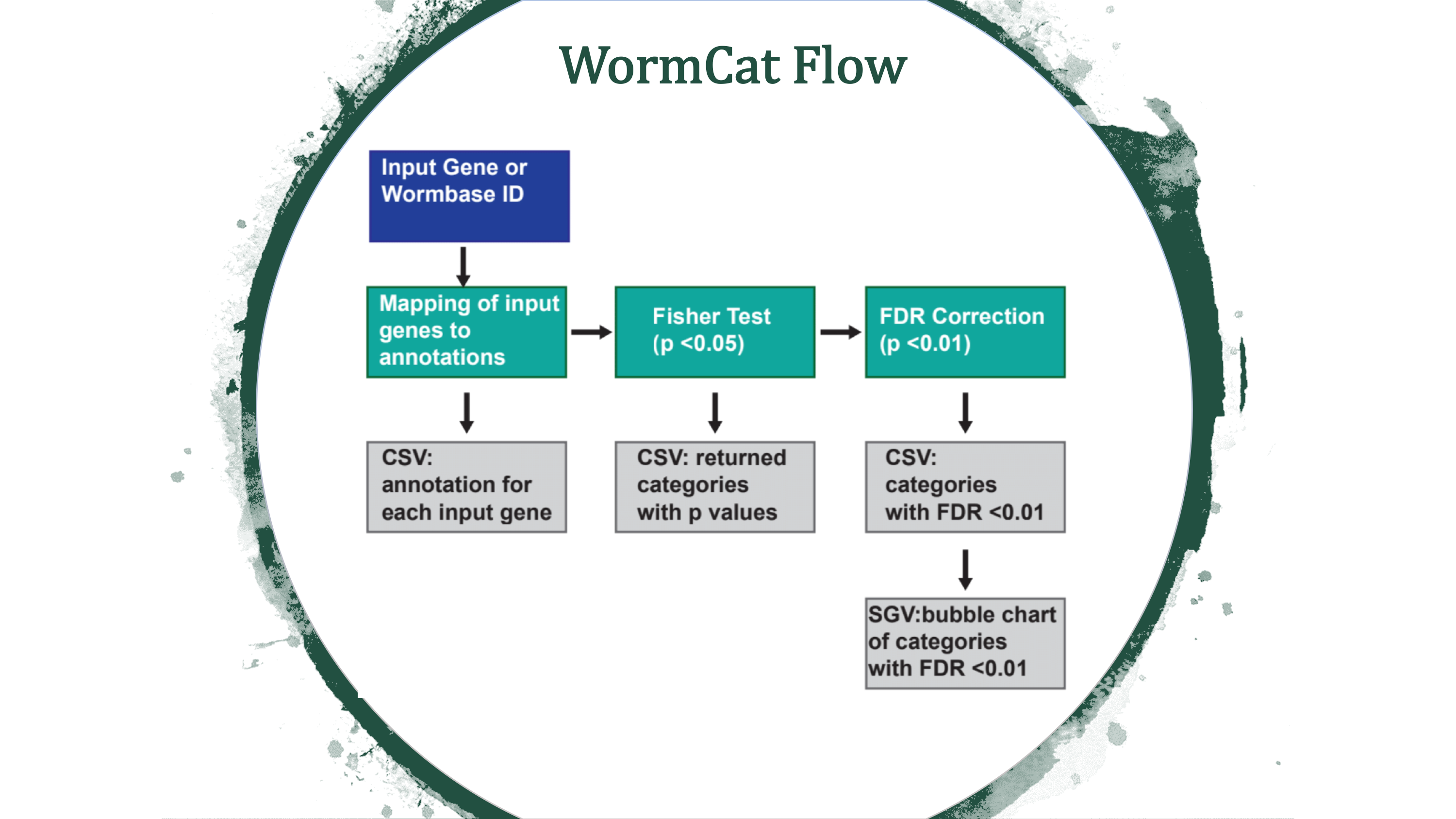
WormCat is a tool for annotating and visualizing gene set enrichment data from C. elegans microarray, RNA seq or RNAi screen data.
- Three Annotation Types are provided
- Whole Genome (1): for microarray or RNA seq data.
- And Orfeome (2) or Ahringer (3) : These are sub-annotation lists and are available for RNAi screen data.
- Two Input Types are allowed
- Sequence ID
- Wormbase ID
- Providing a Search Title will help differentiate results over multiple runs and enhances the output report.
- The Regulated Gene Set data can be loaded either by
- Dropping a file in the Drop File Here zone
- Or pasting data in the Input Box
- Provide us an email and click submit.
A sample file for testing is provided here with Wormbase ID's. (Note: Make sure to select Wormbase ID as the Input Type.)
Output
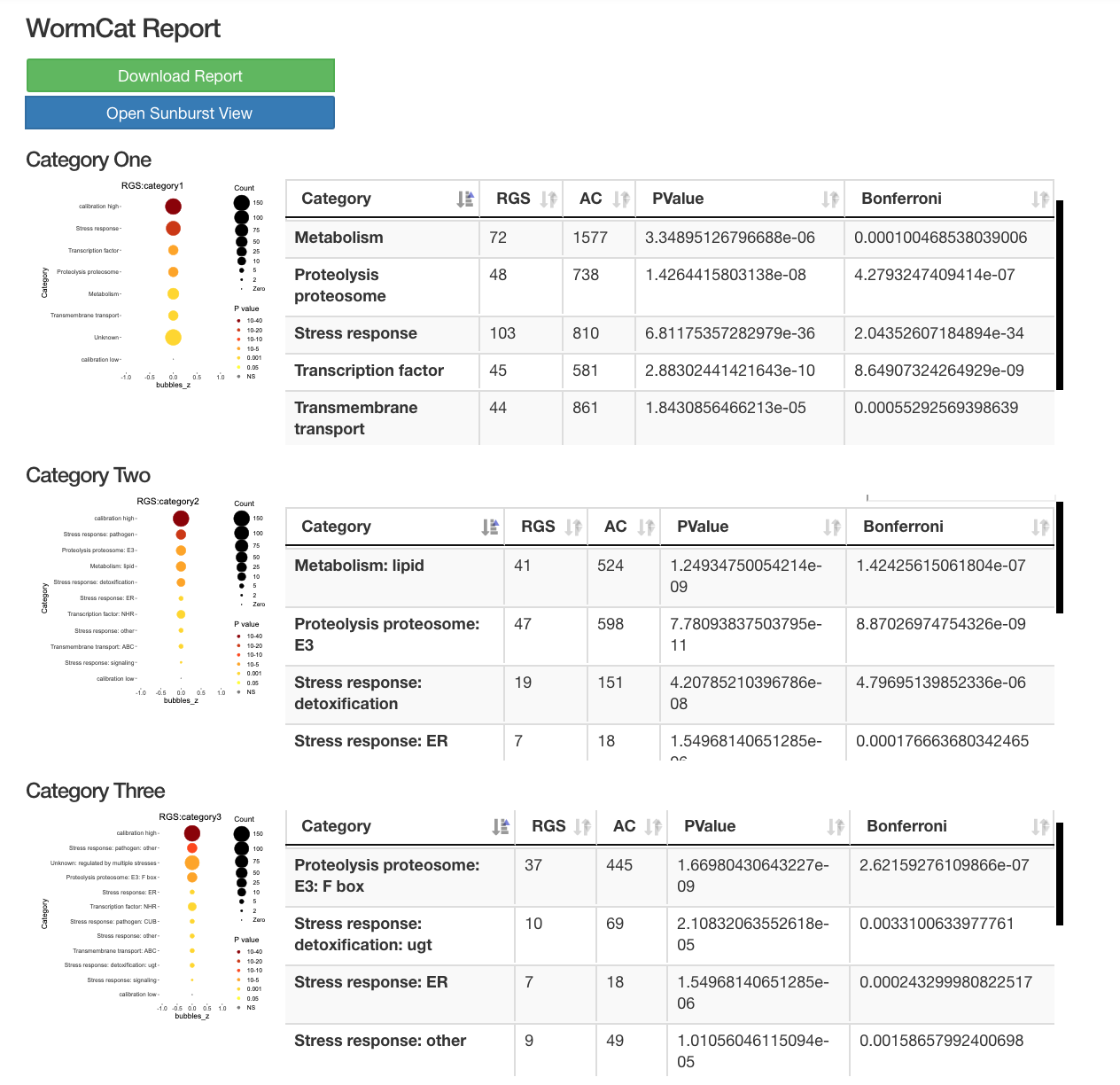
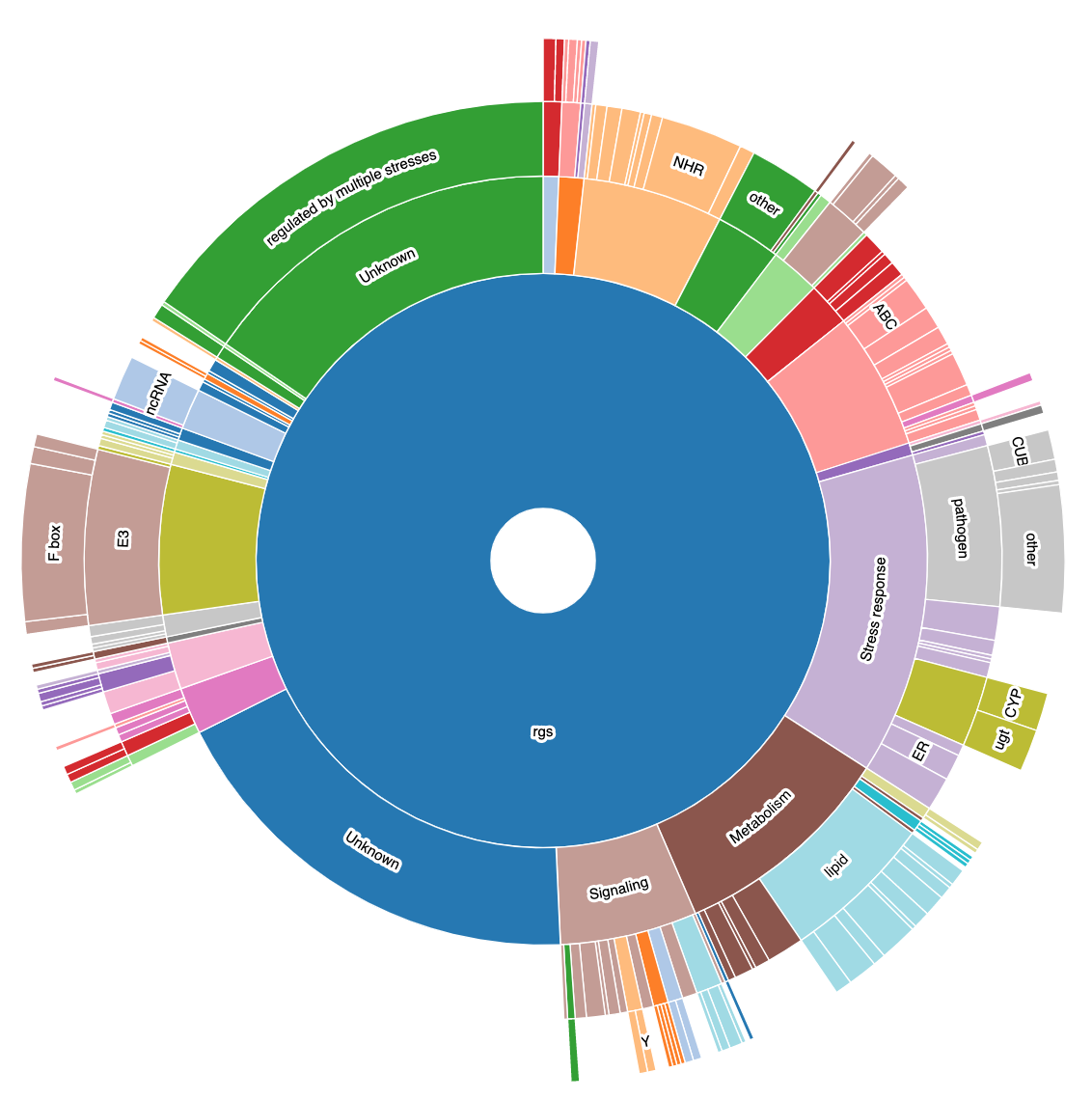
WormCat will provide enrichment data from the nested annotation list with broad categories in Category 1 (Cat1) and more specific categories in Cat2 and Cat3. For example, sbp-1 is in Metabolism: lipid: transcriptional regulator. WormCat output provides scaled bubble charts with enrichment scores that meet a Bonferroni false discovery rate cut off of 0.01 as SGV files. The download directory includes CSV files on the data used for the graph (e.g. rgs_fisher_cat1_apv.csv) (apv is appropriate p value). We also include CSV files with categories that have at least one returned gene and p value from Fisher’s exact test (rgs_fisher_cat1.csv). The rgs_and_category.csv file returns the input gene with annotations.
Multiple Runs
Once you are comfortable running Wormcat with a single dataset, you may wish to execute Wormcat with multiple datasets. Running with multiple datasets is possible by aggregating your datasets into a Microsoft Excel File and dropping the file in the Drop File Here zone.
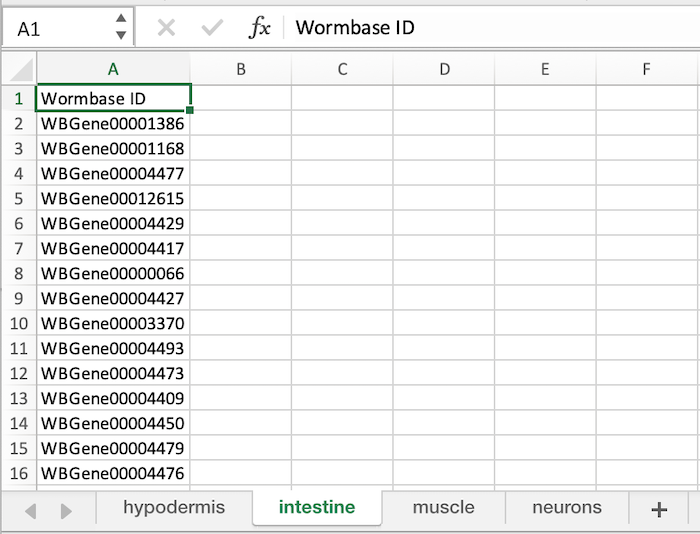
You can download an Example Microsoft Excel file here to confirm the required format. Please take care to follow the naming conventions to avoid processing errors.
- The Spreadsheet Name should ONLY be composed of Letters, Numbers, and Underscores (_) and has an extension .xlsx, .xlt, .xls
- The individual Sheet Names (i.e., Tab name) within the spreadsheet should ONLY be composed of Letters, Numbers, and Underscores (_).
- Each Sheet requires a column header which MUST be 'Sequence ID' or 'Wormbase ID' (The column header is case sensitive.)
Below is an image of a correctly formatted Excel File.
Local Runs
Frequent users might find it useful to install WormCat locally. When running locally a Python script is available on GitHub for batching and running WormCat on multiple gene sets. Additionally when running locally, the annotation list may be modified to accommodate user specific preferences.
We have provided a number of Videos that users may find useful when installing and running Wormcat Locally.
Working with WormCat SVG files in Adobe Illustrator
The SVG files have layers that can be removed to allow the text and circles to be removed for formation of composite WormCat figures.
- Open SVG files in Adobe Illustrator
- If there are a large number of categories or the text is long, the image may appear cropped, the following steps will resolve in this issue
- Illustrator may say the formatting of Tiny is lost, say OK
- Select all
- Remove clipping mask Object: Clipping Mask: Release. Do this 2X
- Individual text or circles should now be able to be selected by clicking directly on them
- Open a new illustrator file, copy and paste circles/text
- At this point, you can arrange the WormCat graphs as best suits your data. In some cases, it’s best to show all the Cat1 data together. In othere cases, you may want to show a specific enrichment category. Cat2/Cat3 data can also be listed under each Cat1. See https://www.genetics.org/content/214/2/279 for examples.
If you use WormCat in a published work please cite:
WormCat: an online tool for annotation and visualization of Caenorhabditis elegans genome-scale data
Amy D Holdorf, Daniel P Higgins, Anne C. Hart, Peter R Boag, Gregory Pazour, Albertha J. M. Walhout, Amy Karol Walker
GENETICS February 1, 2020 vol. 214 no. 2 279-294;